
中国水稻科学 ›› 2017, Vol. 31 ›› Issue (3): 247-256.DOI: 10.16819/j.1001-7216.2017.6159 247
轩丹丹#, 孙廉平#, 张沛沛, 张迎信, 吴玮勋, 杨正福, 占小登, 沈希宏, 曹立勇*( ), 程式华*(
), 程式华*( )
)
收稿日期:2016-12-01
修回日期:2017-02-06
出版日期:2017-05-10
发布日期:2017-05-10
通讯作者:
轩丹丹,孙廉平,曹立勇,程式华
基金资助:
Dandan XUAN#, Lianping SUN#, Peipei ZHANG, Yingxin ZHANG, Weixun WU, Zhengfu YANG, Xiaodeng ZHAN, Xihong SHEN, Liyong CAO*( ), Shihua CHENG*(
), Shihua CHENG*( )
)
Received:2016-12-01
Revised:2017-02-06
Online:2017-05-10
Published:2017-05-10
Contact:
Dandan XUAN, Lianping SUN, Liyong CAO, Shihua CHENG
摘要:
【目的】水稻花药角质层和蜡质层是花粉囊发育重要的结构支撑和安全屏障。对花粉囊发育相关基因进行表型分析和遗传定位,为进一步克隆相关基因和分子机制研究提供基因资源和理论基础。【方法】从籼稻中恢8015辐射诱变(60Co-γ)突变体库中分离鉴定出了一个无花粉型雄性不育突变体whf41。对野生型和突变体不同发育时期的花药进行半薄切片观察;并对其成熟期的花药进行扫描电镜观察。将突变体分别与中恢8015和02428杂交,构建BC1F1、F1株系和BC1F2、F2群体,对该表型进行遗传分析,采用图位克隆的方法精细定位目的基因。【结果】表型分析结果显示,whf41突变体的花药瘦小且呈透明乳白色,花药中不包含花粉粒细胞;半薄切片结果显示,突变体的小孢子无法形成正常的花粉外壁,绒毡层细胞异常膨大而不进行程序性死亡,最终膨胀的绒毡层和花粉细胞碎片逐渐融合并充满药室;扫描电镜结果进一步发现突变体花药内外壁均呈平滑状而缺乏脂类物质,花粉细胞逐渐破碎并降解。遗传分析表明,whf41突变体的无花粉型雄性不育性状受一对隐性核基因控制,我们将该基因精细定位于第3染色体短臂XD-5和XD-11两个标记之间,物理距离45.6 kb,其中包含9个开放阅读框。序列分析显示该区间内细胞色素P450基因LOC_Os03g07250的第4个外显子处存在1个单碱基替换和3个碱基的缺失,导致其翻译序列发生一个氨基酸的替换(天冬氨酸→甲硫氨酸)和一个氨基酸(缬氨酸)的缺失,致使其功能改变进而出现该表型。qRT-PCR检测结果表明,whf41突变体中CYP704B2和一系列花药脂质合成与转运相关基因的表达水平均发生了显著下调。【结论】根据本研究结果可推断,OsWHF41是CYP704B2的新等位基因,相关结果进一步明确CYP704B2在水稻花药脂质合成与花粉壁形成过程中的重要作用。
中图分类号:
轩丹丹, 孙廉平, 张沛沛, 张迎信, 吴玮勋, 杨正福, 占小登, 沈希宏, 曹立勇, 程式华. 水稻无花粉型核雄性不育突变体whf41的鉴定与基因定位[J]. 中国水稻科学, 2017, 31(3): 247-256.
Dandan XUAN, Lianping SUN, Peipei ZHANG, Yingxin ZHANG, Weixun WU, Zhengfu YANG, Xiaodeng ZHAN, Xihong SHEN, Liyong CAO, Shihua CHENG. Characterization and Gene Mapping of a No-pollen Genic Sterile Mutant whf41 in Rice[J]. Chinese Journal OF Rice Science, 2017, 31(3): 247-256.
| 引物名称 Primer name | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') | 实验目的 Purpose | |
|---|---|---|---|---|
| InD40 | CTGCACCGGAGAAATTTGAT | CGCATGCAGATGAATAGGTG | 精细定位 Fine maping | |
| InD41 | TAATTTCGGCTCATCCAAGC | GAAGCTCCGCAGGTTCAG | 精细定位 Fine maping | |
| RM14436 | CTGACGCCGTCTTGCGTTATTCC | CGGCGACTTCGACTACTCAAGC | 精细定位 Fine maping | |
| RM14442 | CGACACGGGCAAGAACTTATACGG | ATCCGATGACGGAGCATGATAGC | 精细定位 Fine maping | |
| XD-5 | TGTCTTGTACCACTGCAATCAT | AACCCAAATCTAACAACTGACG | 精细定位 Fine maping | |
| XD-11 | ACATAAATGATTGTTTCGTGGAG | CCTTTGAGTCAAAAAAAATAGGCA | 精细定位 Fine maping | |
| XD-12 | CACTCAGTGGCAGATCCAAG | GTGCGTGAGTTCGTAAAGATGA | 精细定位 Fine maping | |
| GWHF41 | GAGACGCTCCGCCTCTACC | ACTCCATCCTCCCCATGGA | 突变位点检测 Mutation site detection | |
| LOC_Os06g03930 | CTGCTGTCCCAGTGGATGCTA | CCTCACCCCAAAGGTATGTCA | qRT-PCR | |
| LOC_Os04g48460 | CAAGTTCACGGCGTTCCAG | CGTCATCCCACATCTCGAATCTGAA | qRT-PCR | |
| CYP704B2 | AGCGGCAAGCAAGAGAAGA | GCCATCTTCATCTGGAGGTA | qRT-PCR | |
| CYP703A3 | TTTGATGCAGGACATGATT | TTGCCGGCGCTGAAGGGGA | qRT-PCR | |
| GAMYB | CTCTCCAAAGTTTCCCCAGC | GAACATGATACCGTCGCCAA | qRT-PCR | |
| WDA1 | CAGATCATCCTGAGCGGTAT | CGAGTGGTAGTGGGTGTAGA | qRT-PCR | |
| OSC4 | GAGTGCGTGCCGCAGCTGAA | CTCTCGTCTCGTCTTAGGCAG | qRT-PCR | |
| OSC6 | TGCCACATCGTGAACCACACCCTG | ATGTTGAAATCCCTCCTTTGGTA | qRT-PCR | |
表1 本研究所用引物
Table 1 Primers used in the research.
| 引物名称 Primer name | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') | 实验目的 Purpose | |
|---|---|---|---|---|
| InD40 | CTGCACCGGAGAAATTTGAT | CGCATGCAGATGAATAGGTG | 精细定位 Fine maping | |
| InD41 | TAATTTCGGCTCATCCAAGC | GAAGCTCCGCAGGTTCAG | 精细定位 Fine maping | |
| RM14436 | CTGACGCCGTCTTGCGTTATTCC | CGGCGACTTCGACTACTCAAGC | 精细定位 Fine maping | |
| RM14442 | CGACACGGGCAAGAACTTATACGG | ATCCGATGACGGAGCATGATAGC | 精细定位 Fine maping | |
| XD-5 | TGTCTTGTACCACTGCAATCAT | AACCCAAATCTAACAACTGACG | 精细定位 Fine maping | |
| XD-11 | ACATAAATGATTGTTTCGTGGAG | CCTTTGAGTCAAAAAAAATAGGCA | 精细定位 Fine maping | |
| XD-12 | CACTCAGTGGCAGATCCAAG | GTGCGTGAGTTCGTAAAGATGA | 精细定位 Fine maping | |
| GWHF41 | GAGACGCTCCGCCTCTACC | ACTCCATCCTCCCCATGGA | 突变位点检测 Mutation site detection | |
| LOC_Os06g03930 | CTGCTGTCCCAGTGGATGCTA | CCTCACCCCAAAGGTATGTCA | qRT-PCR | |
| LOC_Os04g48460 | CAAGTTCACGGCGTTCCAG | CGTCATCCCACATCTCGAATCTGAA | qRT-PCR | |
| CYP704B2 | AGCGGCAAGCAAGAGAAGA | GCCATCTTCATCTGGAGGTA | qRT-PCR | |
| CYP703A3 | TTTGATGCAGGACATGATT | TTGCCGGCGCTGAAGGGGA | qRT-PCR | |
| GAMYB | CTCTCCAAAGTTTCCCCAGC | GAACATGATACCGTCGCCAA | qRT-PCR | |
| WDA1 | CAGATCATCCTGAGCGGTAT | CGAGTGGTAGTGGGTGTAGA | qRT-PCR | |
| OSC4 | GAGTGCGTGCCGCAGCTGAA | CTCTCGTCTCGTCTTAGGCAG | qRT-PCR | |
| OSC6 | TGCCACATCGTGAACCACACCCTG | ATGTTGAAATCCCTCCTTTGGTA | qRT-PCR | |
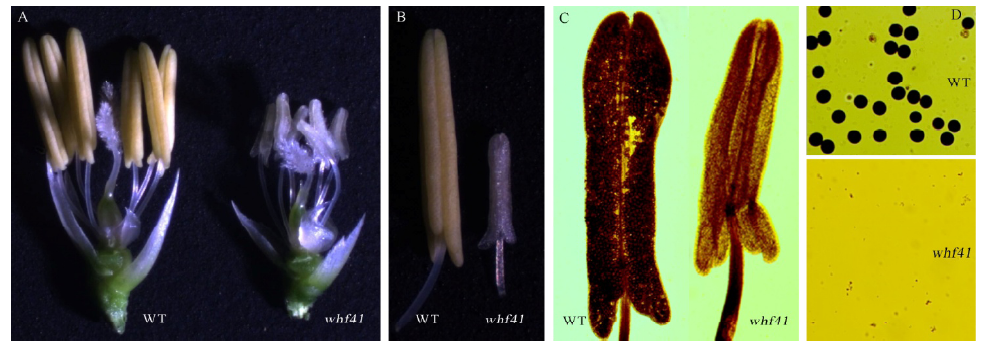
图1 野生型中恢8015(WT)与突变体whf41的表型分析 A–野生型中恢8015和whf41突变体去除内外桴的颖花形态; B–野生型和突变体whf41花药表型; C–野生型和突变体whf41花药压片; D–中恢8015野生型和突变体whf41花粉镜检。
Fig. 1. Phenotypic comparison of wild type Zhonghui 8015(WT) and the whf41 mutant. A, Flower of wild type Zhonghui 8015 and the whf41 mutant with the lemma and palea removed; B, Anther of wild type and the whf41 mutant; C, Anther tablet of wild type and the whf41 mutant; D, I2-KI staining of pollen grains of wild type and the whf41 mutant.
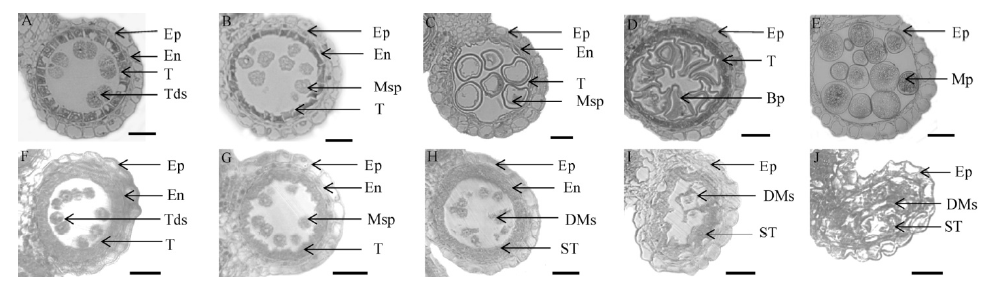
图2 野生型中恢8015和whf41突变体野生型不同发育时期花药半薄横切观察 A到E为野生型,F到J为突变体whf41。A和F–花药发育第8期;B和G–花药发育第9期;C和H–花药发育第10期;D和I–花药发育第11期;E和J–花药发育第12期。 Ep–表皮层;En–内皮层;T–绒毡层;ST–膨胀绒毡层;Tds–四分体;Msp–小孢子;DMs–退化小孢子;Bp–二孢花粉;Mp–成熟花粉;标尺为20 μm。
Fig. 2. Transverse section of anthers of the wild type and whf41 at different developmental stages. A to E, Wild type; F to J, whf41 mutant; A and F, Cross-section of anthers at the stage 8; B and G, Cross-section of anthers at the stage 9; C and H, Cross-section of anthers at the stage 10; D and I, Cross-section of anthers at the stage 11; E and J, Cross-section of anthers at the stage 12. Ep, Epidermis; En, Endothecium; T, Tapetum; ST, Swollen tapetum; Tds, Tetrads; Msp, Microspores; DMs, Degenerated microspores; BP, Biceullar pollen; MP, Mature pollen. Bar=20 μm.
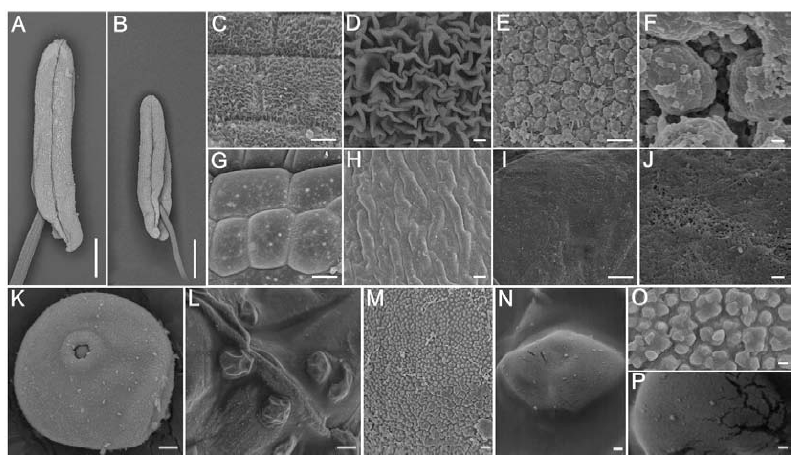
图3 野生型和whf41花药和花粉结构的扫描电镜观察 A和B分别为野生型和whf41第13期的花药;C、D和G、H分别为野生型和whf41第13期的花药外壁;E、F和I、J分别为野生型和whf41第13期的花药绒毡层内表面;K和L分别为野生型和whf41第13期的花粉粒;M、O和N、P分别为野生型和whf41花粉粒第13期的花粉外壁。A和B标尺为20 mm,C~N标尺为5 mm,O和P标尺为200 μm。
Fig. 3. SEM observation of the anther and pollen grains in wild type and the whf41 mutant. A, Anthers at stage 13 of the wild type and the mutant whf41, respectively; C, D and G, H, The outmost surface on epidermis of anthers at stage 13 of the wild type and whf41, respectively. E, F and I, J, The innermost surface on tapetum of anthers at stage 13 of the wild type and whf41, respectively. K and L, The pollen grain of anthers at stage 13 of the wild type and whf41. M, O and N, P, The pollen exine of anthers at stage 13 of the wild type and whf41, respectively. Bar= 20 mm in A and B, 5 mm in C to N, and 200 μm in O and P.
| 组合 Combination | F1结实率 Seed-setting rate of F1/% | F2 | χ2(3:1) | χ20.05 | |
|---|---|---|---|---|---|
| 野生型植株数 No. of wild-type plants | 突变表型植株数 No. of mutant-type plants | ||||
| whf41/中恢8015 whf41/Zhonghui 8015 | 78.73 | 309 | 91 | 1.19 | 3.84 |
| whf41/02428 | 80.51 | 3936 | 1278 | 0.63 | 3.84 |
| 中恢8015/02428 Zhonghui 8015/02428 | 85.98 | 6648 | 0 | - | 3.84 |
表2 whf41突变位点的遗传分析
Table 2 Genetic analysis of the whf41 locus.
| 组合 Combination | F1结实率 Seed-setting rate of F1/% | F2 | χ2(3:1) | χ20.05 | |
|---|---|---|---|---|---|
| 野生型植株数 No. of wild-type plants | 突变表型植株数 No. of mutant-type plants | ||||
| whf41/中恢8015 whf41/Zhonghui 8015 | 78.73 | 309 | 91 | 1.19 | 3.84 |
| whf41/02428 | 80.51 | 3936 | 1278 | 0.63 | 3.84 |
| 中恢8015/02428 Zhonghui 8015/02428 | 85.98 | 6648 | 0 | - | 3.84 |

图4 突变基因的图位克隆 A–突变基因的初步定位;B–突变基因的精细定位;C–精细定位区间内的开放阅读框;D–候选基因LOC_Os03g07250结构;E–whf41突变位点及蛋白序列。
Fig. 4. Positional cloning of the whf41 mutated gene. A, Preliminary mapping of whf41; B, Fine mapping of whf41; C, The ORFs within the fine-mapped region; D, The structure candidate gene LOC_Os03g07250; E, The mutation site and translation sequence in whf41 mutant.
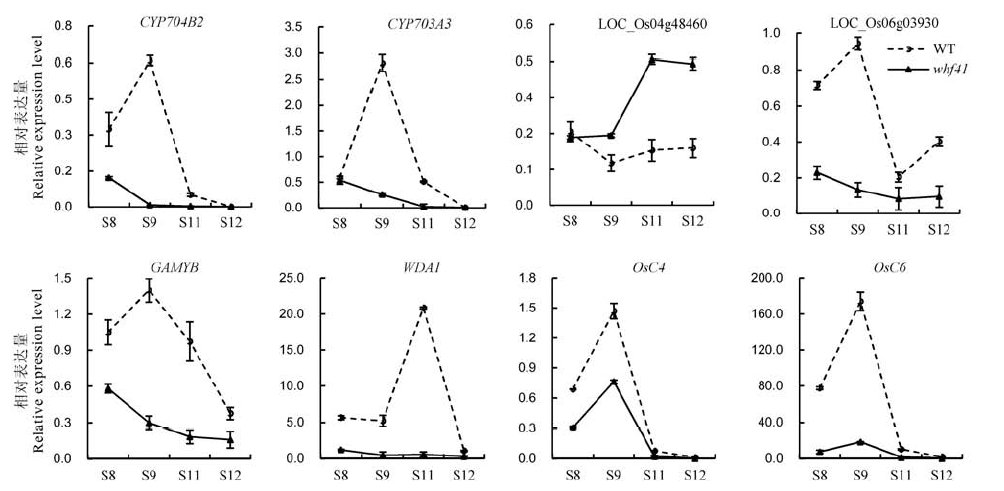
图5 野生型与whf41 突变体中水稻花药发育相关基因的表达以OsActin1作为对照,误差线表示n=3时的标准误。S8,S9,S11和S12分别代表花药发育的第8、9、11和12期;WT–野生型。
Fig. 5. Expression profile of the genes involved in anther development in wild type and whf41 plants. OsActin1 was used as an internal control. Error bars show the SD (n = 3). S8, S9, S11, and S12 indicated stage 8, stage 9, stage 11 and stage 12 of anther development; WT, Wild type.
| [1] | 朱英国. 水稻雄性不育生物学. 武汉: 武汉大学出版社, 2000: 1-2. |
| Zhu Y G.Biology of Male Sterility in Rice. Wuhan: Wuhan University Press, 2000: 1-2. (in Chinese) | |
| [2] | Raghavan V.Anther developmental biology in molecular embryology of flowering plants.Plant Physiol, 1997: 17-60. |
| [3] | Ma H.Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants.Annu Rev Plant Boil, 2005, 56: 393-434. |
| [4] | Zhang D B, Wilson Z A.Stamen specification and anther development in rice. Chin Sci Bull, 2009, 54: 2342-2353. |
| [5] | Zhang D B, Luo X, Zhu L.Cytological analysis and genetic control of rice anther development.J Gen Genom, 2011, 38(9): 379-390. |
| [6] | Kinoshita T.Gene symbols and information on male sterility.Rice Genet Newsl, 1997, 14: 13-22. |
| [7] | Okamuro J K, Boer B G, Jofuku K D.Regulation of Arabidopsis flower development.Plant Cell, 1993, 5: 1183-1193. |
| [8] | 马西青, 方才臣, 邓联武, 万向元. 水稻隐性核雄性不育研究进展及育种应用探讨.中国水稻科学,2012, 25(5): 511-520. |
| Ma X Q, Fang C C, Deng L W, Wan X Y.Research progress and breeding application of recessive genic male sterility gene in rice.Chin J Rice Sci, 2012, 25(5): 511-520. (in Chinese with English abstract) | |
| [9] | Nonomura K I, Miyoshi K, Eiguchi M, Suzuki T, Miyao A, Hirochika H, Kurata N.The MSP1 gene is necessary to restrict the number of cells entering into male and female sporogenesis and to initiate anther wall formation in rice.Plant Cell, 2003, 15(8): 1728-1739. |
| [10] | Nonomura K I, Nakano M, Fukuda T, Eiguchi M, Miyao A, Hirochika H, Kurata N.The novel gene HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 of rice encodes a putative coiled-coil protein required for homologous chromosome pairing in meiosis.Plant J, 2004, 16(4): 1008-1020. |
| [11] | Nonomura K I, Nakano M, Murata K, Miyoshi K, Eiguchi M, Miyao A, Hirochika H, Kurata N.An insertional mutation in the rice PAIR2 gene, the ortholog of Arabidopsis ASY1, results in a defect in homologous chromosome pairing during meiosis.Mol Gen Genom, 2004, 271(2): 121-129. |
| [12] | Yuan W Y, Li X W, Chang Y X, Wen R Y, Chen G X, Zhang Q F, Wu C.Mutation of the rice gene PAIR3 results in lack of bivalent formation in meiosis.Plant J, 2009, 59(2): 303-315. |
| [13] | Wang M, Wang K J, Tang D, Wei C X, Li M, Shen Y, Chi Z C, Gu M H, Cheng Z K.The central element protein ZEP1 of the synaptonemal complex regulates the number of crossovers during meiosis in rice.Plant Cell, 2010, 22: 417-430. |
| [14] | Nonomura K, Morohoshi A, Nakano M, Eiguchi M, Miyao A, Hirochika H, Kurata N.A germ cell-specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice.Plant Cell, 2007, 19(8): 2583-2594. |
| [15] | Wang C, Yue W, Ying Y, Wang S, Secco D, Liu Y, Whelan J, Tyerman S D, Shou H.Rice SPX-Major facility superfamily3, a vacuolar phosphate efflux transporter, is involved in maintaining phosphate homeostasis in rice.Plant Physiol, 2015, 169(4): 2822-2831. |
| [16] | He Y, Wang C, Higgins J D, Yu J, Zong J, Lu P, Zhang D, Liang W.MEIOTIC F-BOX is essential for male meiotic DNA double-strand break repair in rice.Plant Cell, 2016, 28(8): 1879-1893. |
| [17] | Li L, Li Y, Song S, Deng H, Li N, Fu X, Chen G, Yuan L.An anther development F-box (ADF) protein regulated by tapetum degeneration retardation (TDR) controls rice anther development.Planta, 2015, 241(1): 157-166. |
| [18] | Jung K H, Han M J, Lee Y S, Kim Y W, Hwang I, Kim M J, Kim Y K, Nahm B H, An G.Rice Undeveloped Tapetum1 is a major regulator of early tapetum development.Plant Cell, 2005, 17(10): 2705-2722. |
| [19] | Liu Z, Bao W, Liang W, Yin J, Zhang D.Identification of gamyb-4 and analysis of the regulatory role of GAMYB in rice anther development.J Integr Plant Biol, 2010, 52(7): 670-678. |
| [20] | Li H, Yuan Z, Vizcay-Barrena G, Yang C, Liang W, Zong J, Wilson Z A, Zhang D.PERSISTENT TAPETAL CELL1 encodes a PHD-Finger protein that is required for tapetal cell death and pollen development in rice.Plant Physiol, 2011, 156(2): 615-630. |
| [21] | Li X, Gao X, Wei Y, Deng L, Ouyang Y, Chen G, Li X, Zhang Q, Wu C.Rice APOPTOSIS INHIBITOR5 coupled with two DEAD-box adenosine 5′-triphosphate- dependent RNA helicases regulates tapetum degeneration.Plant Cell, 2011, 23(4): 1416-1434. |
| [22] | Niu N, Liang W, Yang X, Jin W, Wilson Z A, Hu J, Zhang D.EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat Comm, 2013, 4: 1445. |
| [23] | Fu Z, Yu J, Cheng X, Zong X, Xu J, Chen M, Li Z, Zhang D, Liang W.The rice basic Helix-Loop-Helix transcription factor TDR INTERACTING PROTEIN2 is a central switch in early anther development.Plant Cell, 2014, 26(4): 1512-1524. |
| [24] | Niu B X, He F R, He M, Ren D, Chen L T, Liu Y G.The ATP-binding cassette transporter OsABCG15 is required for anther development and pollen fertility in rice.J Integr Plant Biol, 2013, 55(8): 710-720. |
| [25] | Zhao G, Shi J, Liang W, Xue F, Luo Q, Zhu L, Qu G, Chen M, Schreiber L, Zhang D.Two ATP binding cassette G transporters, rice ATP binding cassette G26 and ATP binding cassette G15, collaboratively regulate rice male reproduction.Plant Physiol, 2015, 169(3): 2064-2079. |
| [26] | Zhang D, Liang W, Yin C, Zong J, Gu F, Zhang D.OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice.Plant Physiol, 2010, 154(1): 149-162. |
| [27] | Jung K H, Han M J, Lee D Y, Lee Y S, Schreiber L, Franke R, Faust A, Yephremov A, Saedler H, Kim Y W, Hwang I, An G.Wax-Deficient Anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development.Plant Cell, 2006, 18(11): 3015-3032. |
| [28] | Shi J, Tan H, Yu X H, Liu Y, Liang W, Ranathunge K, Franke R B, Schreiber L, Wang Y, Kai G, Shanklin J, Ma H, Zhang D.Defective Pollen Wall is required for anther and microspore development in rice and encodes a fatty acyl carrier protein reductase.Plant Cell, 2011, 23(6): 2225-2246. |
| [29] | Xu D W, Shi J X, Rautengarten C, Yang L, Qian X L, Uzair M, Zhu L, Luo Q, An G, Waßmann F, Schreiber L, Heazlewood J L, Scheller H V, Hu J P, Zhang D B, Liang W Q.Defective Pollen Wall 2 (DPW2) encodes an acyl transferase required for rice pollen development.Plant Physiol, 2017, 173(1): 240-255. |
| [30] | Li H, Pinot F, Sauveplane V, Werck-Reichhart D, Diehl P, Schreiber L, Franke R, Zhang P, Chen L, Gao Y, Liang W, Zhang D.Cytochrome P450 family member CYP704B2 catalyzes the ω -hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell, 2010, 22(1): 173-190. |
| [31] | Yang X, Wu D, Shi J, He Y, Pinot F, Grausem B, Yin C, Zhu L, Chen M, Luo Z, Liang W, Zhang D.Rice CYP703A3, a cytochrome P450 hydroxylase, is essential for development of anther cuticle and pollen exine.J Integr Plant Biol, 2014, 56(10): 979-994. |
| [32] | Zhu Q H, Ramm K, Shivakkumar R, Dennis E S, Upadhyaya N M.The ANTHER INDEHISCENCE1 gene encoding a single MYB domain protein is involved in anther development in rice.Plant Physiol, 2004, 135(3): 1514-1525. |
| [33] | Kent B, Erin C, Nina D, David H, Young J K, Zhong Cathy X Y. Plant genomic DNA flanking SPT event and methods for identifying SPT event. United States Patent, US008257930B2, 20120904. |
| [34] | Chang Z Y, Chen Z F, Wang N, Xie G, Lu J W, Yan W, Zhou J L, Tang X Y, Deng X W.Construction of a male sterility system for hybrid rice breeding and seed production using a nuclear male sterility gene.Proc Natl Acad Sci USA, 2016, 113(49): 14145. |
| [35] | Sun L P, Zhang Y X, Zhang P P, Yang Z F, Zhan X D, Shen X H, Zhang Z H, Hu X, Xuan D D, Wu W X, Li Z H, Cao L Y, Cheng S H.K-Domain splicing factor OsMADS1 regulates open hull male sterility in rice.Rice Sci, 2015, 22(5): 207-216. |
| [36] | Orjuela J, Garavito A, Bouniol M, Arbelaez J D, Moreno L, Kimball J, Wilson G, Rami J F, Tohme J, McCouch S R, Lorieux M. A universal core genetic map for rice.Theor Appl Genet, 2010, 120: 563-572 |
| [37] | Wu D H, Wu H P, Wang C S, Tseng H Y, Hwu K K.Genome-wide InDel marker system for application in rice breeding and mapping studies.Euphytica, 2013, 192: 131-143 |
| [38] | Yamanaka S, Suzuki E, Tanaka M, Takeda Y, Watanabe J A, Watanabe K N.Assessment of cytochrome P450 sequences offers a useful tool for determining genetic diversity in higher plant species.Theor Appl Genet, 2003, 108(1): 1-9. |
| [39] | Tang W, Wu T, Ye J, Sun J, Jiang Y, Yu J, Tang J, Chen G, Wang C, Wan J.SNP-based analysis of genetic diversity reveals important alleles associated with seed size in rice.BMC Plant Biol, 2016, 16(1): 93. |
| [1] | 郭展, 张运波. 水稻对干旱胁迫的生理生化响应及分子调控研究进展[J]. 中国水稻科学, 2024, 38(4): 335-349. |
| [2] | 韦还和, 马唯一, 左博源, 汪璐璐, 朱旺, 耿孝宇, 张翔, 孟天瑶, 陈英龙, 高平磊, 许轲, 霍中洋, 戴其根. 盐、干旱及其复合胁迫对水稻产量和品质形成影响的研究进展[J]. 中国水稻科学, 2024, 38(4): 350-363. |
| [3] | 许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374. |
| [4] | 候小琴, 王莹, 余贝, 符卫蒙, 奉保华, 沈煜潮, 谢杭军, 王焕然, 许用强, 武志海, 王建军, 陶龙兴, 符冠富. 黄腐酸钾提高水稻秧苗耐盐性的作用途径分析[J]. 中国水稻科学, 2024, 38(4): 409-421. |
| [5] | 胡继杰, 胡志华, 张均华, 曹小闯, 金千瑜, 章志远, 朱练峰. 根际饱和溶解氧对水稻分蘖期光合及生长特性的影响[J]. 中国水稻科学, 2024, 38(4): 437-446. |
| [6] | 刘福祥, 甄浩洋, 彭焕, 郑刘春, 彭德良, 文艳华. 广东省水稻孢囊线虫病调查与鉴定[J]. 中国水稻科学, 2024, 38(4): 456-461. |
| [7] | 陈浩田, 秦缘, 钟笑涵, 林晨语, 秦竞航, 杨建昌, 张伟杨. 水稻根系和土壤性状与稻田甲烷排放关系的研究进展[J]. 中国水稻科学, 2024, 38(3): 233-245. |
| [8] | 缪军, 冉金晖, 徐梦彬, 卜柳冰, 王平, 梁国华, 周勇. 过量表达异三聚体G蛋白γ亚基基因RGG2提高水稻抗旱性[J]. 中国水稻科学, 2024, 38(3): 246-255. |
| [9] | 尹潇潇, 张芷菡, 颜绣莲, 廖蓉, 杨思葭, 郭岱铭, 樊晶, 赵志学, 王文明. 多个稻曲病菌效应因子的信号肽验证和表达分析[J]. 中国水稻科学, 2024, 38(3): 256-265. |
| [10] | 朱裕敬, 桂金鑫, 龚成云, 罗新阳, 石居斌, 张海清, 贺记外. 全基因组关联分析定位水稻分蘖角度QTL[J]. 中国水稻科学, 2024, 38(3): 266-276. |
| [11] | 魏倩倩, 汪玉磊, 孔海民, 徐青山, 颜玉莲, 潘林, 迟春欣, 孔亚丽, 田文昊, 朱练峰, 曹小闯, 张均华, 朱春权. 信号分子硫化氢参与硫肥缓解铝对水稻生长抑制作用的机制[J]. 中国水稻科学, 2024, 38(3): 290-302. |
| [12] | 周甜, 吴少华, 康建宏, 吴宏亮, 杨生龙, 王星强, 李昱, 黄玉峰. 不同种植模式对水稻籽粒淀粉含量及淀粉关键酶活性的影响[J]. 中国水稻科学, 2024, 38(3): 303-315. |
| [13] | 关雅琪, 鄂志国, 王磊, 申红芳. 影响中国水稻生产环节外包发展因素的实证研究:基于群体效应视角[J]. 中国水稻科学, 2024, 38(3): 324-334. |
| [14] | 许用强, 姜宁, 奉保华, 肖晶晶, 陶龙兴, 符冠富. 水稻开花期高温热害响应机理及其调控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 111-126. |
| [15] | 吕海涛, 李建忠, 鲁艳辉, 徐红星, 郑许松, 吕仲贤. 稻田福寿螺的发生、危害及其防控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 127-139. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||